How to complete the quote request form
Project submission process
1. Quote request form
Download, complete and send to igf@imperial.ac.uk.
2. Quotation
Sign and return the quotation received (ask for any changes needed before signing).
3. Sample submission form
Complete and return prior to submitting the samples to the Facility. Information on the “Container ID” column must exactly match the labels on your individual tubes/plates.
If you requested data analysis, we will need you to provide additional information regarding the experimental design, metadata and sample grouping. For this quote, we assume that each sample is only used once for data analysis, should any sample be needed more than once (e.g. control vs group1 and control vs group2) the cost for analysis will be adjusted by counting the control sample(s) twice.
4. Sample drop-off
- Prepare and deliver: samples/libraries to the facility in accordance with Sample Submission Guidelines (Further information below).
- QC Plate: a separate labelled QC plate is required with at least 4μl of each sample.
- Containers: samples/libraries must be submitted in 96-well plates, with samples loaded in columns.
- Label: plates with the Container ID, Quotation ID, the PI name and the date.
- An individual container ID needed on the form per each submitted plate.
- We cannot accept samples/libraries/data prior to the receipt of a sample submission form.
5. Project starts
- Initial QC: Your samples/libraries/data will be initially quality checked (QC) to make sure that the quality is acceptable for the requested application/service.
- Invoice: You will be invoiced according to the agreed quotation or any approved changes to your project.
- Result: You will receive results once your samples/libraries/data have been processed and the invoice has been paid.
Sample submission guidelines
Quantify libraries by qPCR and adjust the concentration of library or pool to 4-10nM by using Tris-Cl 10mM, pH 8.5 with 0.1% Tween 20.
Single library per lane:
- Submit 15 uL of 4-10 nM library
More than one library per lane(s):
- Submit 15 uL of 4-10 nM final pool per lane of sequencing requested
Prior to pooling barcoded libraries, it is essential to normalize the molar concentration of the libraries to ensure that an equal number of reads is generated for each library. In order to Convert ng/µl to nM and normalize library concentrations for pooling:
1. Determine the concentration (ng/µl) of each individual library using Qubit or KAPA Library Quantification Kits.
2. Determine the average size (bp) of each individual library using an Agilent Technologies 2100 Bioanalyzer.
3. Determine the molarity (nM) of each individual library using the figures above and the formula below:
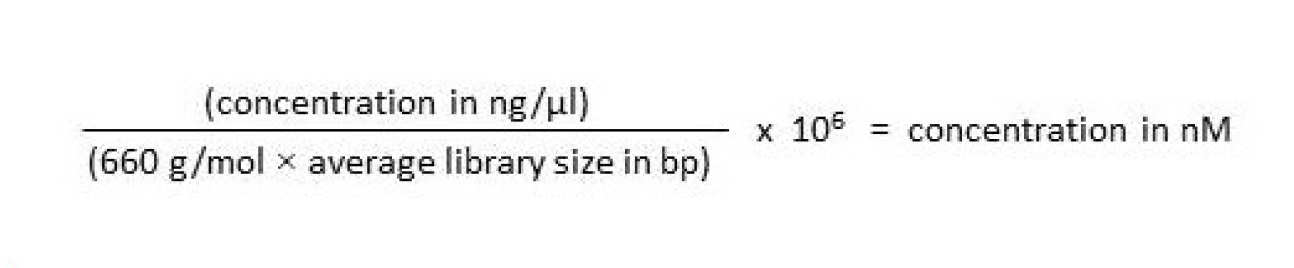
4. Plan your library normalization (dilution calculations): Determine a common concentration (4-10nM) in order to dilute all individual libraries. Dilutions should be done using Tris-Cl 10mM, pH 8.5 with 0.1% Tween 20. Calculate the dilution of each individual library using the equation below:

5. Pool the normalized libraries: Combine equal volumes of each normalized library into a 1.5ml Eppendorf tube and gently pipette contents up and down 10 times to mix thoroughly. submit 15 µL of the pool per lane of sequencing requested.
6. The tube labels must exactly match the “Container ID” that you enter on the sample submission form. An individual ID must per provided per each tube or plate.
*Note: If the library pool concentration is below the submission concentrations mentioned above, contact IGF Facility for further instructions and advice.
Custom sequencing primers
If using custom sequencing primers, these must be submitted along with samples.
- Aliquots must be at 100 uM in at least 20 ul
- Primers must be diluted in EB or DI Water
- Must be in a Low-bind tube and clearly labelled
Important
- All libraries will be stored by the IGF Facility only for three months and then disposed of.
- Please contact us if you wish to collect your library after sequencing. We recommend submission of only an aliquote of your total library prep.
- Libraries received without the appropriate and correct paperwork or poorly labelled, will delay the initiation of your project, risk the safety of your samples and incur additional charges.
1. RNA Extraction:
Total RNA can be extracted using common RNA-extraction kits such as Qiagen RNAeasy kits, Zymo kits (Direct-zol or Quick-RNA miniprep kits) or Invitrogen TRIzol.
2. DNase Treatment:
DNase I treatment needs to be included in the RNA-extraction in order to eliminate the background genomic DNA contamination. Zymo include DNase in their extraction kits or DNase should be purchased separately and added in extraction (TURBO DNase, ThermoFisher).
3. RNA Quality and Volume:
Provide RNA in minimum 13uL RNase-free water (not in TE or other buffers). Our current protocol requires an input amount of 10 ng–1 µg total RNA quantified by Qubit Fluorometer. However, we recommend providing higher amounts (min 300ng). High- quality RNA is essential for successful RNA-Seq experiments. High-quality RNA shows a 28S rRNA band at 4.5 kb of twice the intensity of the 18S rRNA band at 1.9 kb. RNA with a RIN value above 7.
If high-quality RNA is not available, our Facility has access to specific library preparation kits that are designed for partially-degraded and FFPE samples with low RIN values. However, the effect of degraded RNA on the sequencing results should be carefully considered.
4. RNA storage:
RNase-free conditions must be used in handling the RNA samples. In order to minimize the freeze and thaw cycles of your RNA, please please transfer a 4uL aliquot of your extracted samples in separate PCR strip tubes to be used for the initial QC in our Facility. tore both original tubes/plates and QC aliquots at -80C.
5. Dry ice shipment only.
Important:
Following the QC of your RNA samples, we will proceed with samples that meet the minimum requirements for the chosen kit. If any samples fails our QC, we'll contact you and give you the option to proceed, remove the sample(s) from the batch for library prep or consider alternative library construction methods. The project and final invoice will be changed accordingly.
1. DNA Extraction:
Genomic DNA should be purified using a column-based purification protocol such as the Qiagen DNeasy kit, or one of the DNA purification kits from Zymo Research (Quick-gDNA MiniPrep, Quick-gDNA Blood MiniPrep, or ZR Genomic DNA-Tissue MiniPrep). We recommend elution of the DNA in DNase-free water or tris buffer that does not have EDTA as it may interfere with enzyme activities during the library preparation.
2. RNase Treatment:
RNase treatment needs to be included in the DNA-extraction in order to eliminate the background RNA contamination.
3. DNA Quality Check:
Genomic DNA should be provided in DNase-free water as high molecular weight DNA with no RNA contamination. If samples have been exposed to phenol or other organic solvents, they should be run through a Qiagen cleanup column prior to submission to avoid contaminants that may inhibit the activities of enzymes used in the Illumina library preparation protocols. DNA can be stored at -20C.
4. Volume and concentration requirements:
This can vary and is dependent on the kit that is being used.
5. DNA storage recommendations:
Extracted DNA can be stored at -20C.
6. Ice or dry ice shipment
7. Samples must be submitted in clearly labelled 96-well PCR plates arranged in columns.
Important:
Following the QC of your DNA samples, we will proceed with samples that meet the minimum requirements for the chosen kit. If any samples fail our QC, we'll contact you and give you the option to proceed, remove the sample(s) from the batch for library prep or consider alternative library construction methods. The project and final invoice will be changed accordingly.
We can only process material which has already been immunoprecipitated by the laboratory requesting service.
1. Chromatin fragmentation done by you:
We recommend optimising your chromatin fragmentation conditions before performing immunoprecipitation in order to maximise the amount of material in the range of 50 to 350bp.
2. Quality check done by you:
A test for enrichment can be performed by qPCR for regions expected to be bound by the protein of interest. Beware that if chromatin fragmentation is suboptimal and produces predominantly large fragments, the qPCR enrichment test may still be positive but sequencing data will possibly fail to reveal the same enrichment. This is because large fragments are very inefficient or completely fail to generate good quality clusters and will not contribute to the set of sequences.
Quantification should be done using Qubit or picogreen as nanodrop overestimates the amount of material present.
3. Immunoprecipitated DNA concentration and volume:
We require 10ng of material. Samples should be in a volume of no more than 20μl of ultrapure water. Input samples may need dilution, however, please do not dilute
ChIP samples as this may make them undetectable when we perform our QC of your samples.
4. DNA storage recommendations:
5. You can submit your DNA samples on ice.
6. Samples must be submitted in clearly labelled 96-well PCR plates arranged in columns.
7. Important:
Immunoprecipitated samples are often very diluted and may be undetectable when we perform our QC, however, we will proceed with library prep of all samples. Following the QC of the libraries, if any samples fail our QC, we'll contact you and give you the option to proceed or remove the sample(s) from the batch for sequencing. In either case, we will charge for library construction.
More detailed information regarding experimental design and analysis is provided by Landt et al.
