Introduction
Antithrombin is a plasma inhibitor of thrombin and other blood coagulation proteinases. Its (functional) deficiency is a strong risk factor for venous thrombosis. The gene coding for antithrombin has been localised to chromosome 1q23-25. The nucleotide sequence is available from the US National Center for Biotechnology Information (NCBI). A single copy has seven exons spanning 13.4 kb of DNA. Within the introns of the gene are located nine full length and one partial Alu repeat elements (jump to figures I & II)
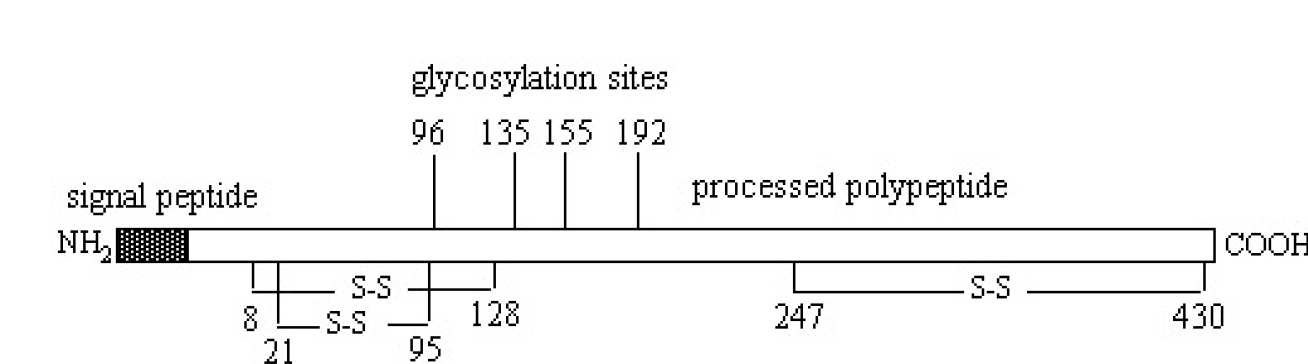
 Two of these Alu5 and Alu8 within intron 4 have tails composed of ATT trinucleotide repeats that are polymorphic in copy number (for gene sequence of anthrombin, see Olds et al, 1993). The gene codes for a protein of MW ~58 000 containing 432 amino acids, 6 of which are cysteines that form three intramolecular disulphide bonds. Antithrombin is a member of a superfamily of proteins collectively known as serine proteinase inhibitors (serpins). Most of the members of this family are inhibitors that control proteolysis of key blood cellular and tissue enzymes. The crystal structures of several members of this family have been determined and these structures have aided understanding of the mechanism of action of the serpins, including antithrombin (for a recent review of serpin structures, see Whisstock et al,1998). Antithrombin contains a functional reactive site which participates in the inhibitory interaction with proteinases, but also a binding site for heparin and related glycosaminoglycans. Binding of heparin to the latter site induces a conformational change and accelerates its inhibition of proteinases (see for example Desai et al, 1998; Huntington & Gettins, 1998). The nature of the heparin binding site on antithrombin has been investigated initially by chemical modification, by investigation of natural and recombinant mutants, and recently by high resolution crystallography (see Jin et al, 1997).
Two of these Alu5 and Alu8 within intron 4 have tails composed of ATT trinucleotide repeats that are polymorphic in copy number (for gene sequence of anthrombin, see Olds et al, 1993). The gene codes for a protein of MW ~58 000 containing 432 amino acids, 6 of which are cysteines that form three intramolecular disulphide bonds. Antithrombin is a member of a superfamily of proteins collectively known as serine proteinase inhibitors (serpins). Most of the members of this family are inhibitors that control proteolysis of key blood cellular and tissue enzymes. The crystal structures of several members of this family have been determined and these structures have aided understanding of the mechanism of action of the serpins, including antithrombin (for a recent review of serpin structures, see Whisstock et al,1998). Antithrombin contains a functional reactive site which participates in the inhibitory interaction with proteinases, but also a binding site for heparin and related glycosaminoglycans. Binding of heparin to the latter site induces a conformational change and accelerates its inhibition of proteinases (see for example Desai et al, 1998; Huntington & Gettins, 1998). The nature of the heparin binding site on antithrombin has been investigated initially by chemical modification, by investigation of natural and recombinant mutants, and recently by high resolution crystallography (see Jin et al, 1997).
The natural mutants of antithrombin, mostly identified from families with a tendency towards thrombosis, are collected together in this database which has been published conventionally (Lane et al, 1991) and recently updated (Lane et al, 1993; Lane et al, 1997). This database is compiled by members of the Plasma Coagulation Inhibitors Subcommittee of the Scientific and Standardization Committee of the International Society on Thrombosis and Haemostasis (SSC of the ISTH). The final version of the database was put together by Trevor Bayston and David Lane. It was reformatted for the web by Malcolm Duncan (Medical Object Oriented Software Enterprises) and Helen Philippou, David Lane (Centre for Haematology, Department of Medicine, Imperial College).
The naming convention accords with "Recommendations for a Nomenclature System for Human Gene Mutations" (see Antonarakis et al, 1998), except that the nucleotide numbering of the original database has been retained (see Olds et al,1993 and Lane et al, 1997). Each entry has been assigned a unique identifier on the basis of the subclass and position in the table for its subclass (e.g. type I case 11 has the mutation 2606insT). The identified mutations are grouped according to a classification system proposed (Lane et al, 1992) and accepted by the SSC of the ISTH. In this, type I and type II deficiencies are distinguished, largely by the presence of variant protein in the latter. Then type II deficiency is further subclassified on the basis of mutations that alter the function of the reactive site, the heparin binding site, and those that have multiple or pleiotropic effects. The database will be periodically updated, depending on the rate at which new mutations are identified. To have mutations included, send information in the format given in the tables to Professor David Lane at d.lane@imperial.ac.uk.
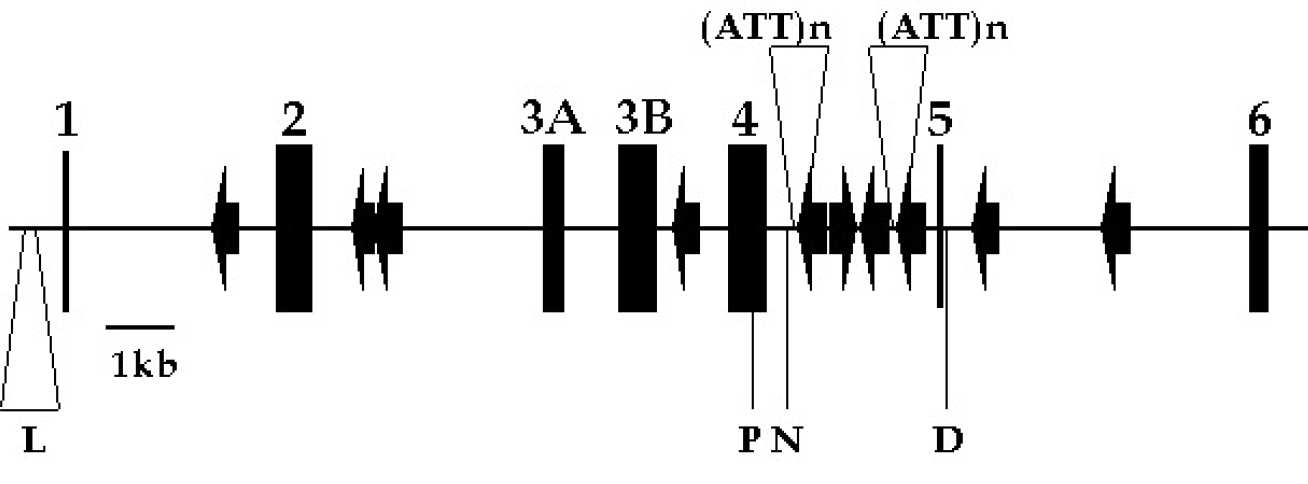
Figures I & II - Structural organisation of the antithrombin gene and protein
The gene structure illustrates the coding regions (exons, numbered 1-6), the Alu repeat regions (arrow heads), and several of the polymorphic sites (L, the 76bp dimorphism at -276; P, Pst I site at 7626; N, the Nhe I site at 7987; D, Dde I site at 9893; (ATT)n, the two trinucleotide polymorphic sites at 8136 and 9289.
Database
- Type I (Classical) deficiency: Point mutations, small (less than 30 bp) insertions and deletions
- Type I (Classical) deficiency: Partial (greater than 30 bp) and whole gene deletions
- Type II deficiency: Heparin binding defects
- Type II deficiency: Pleiotropic effects
- Type II deficiency: Reactive site defects
- Polymorphisms/sequence variations
- Nature of mutations in antithrombin deficiency
- Antithrombin gene mutation by subtype
- CpG dinucleotide mutations in antithrombin deficiency
- References
- Acknowledgements and credits
| New | Nucleotide Position, Mutation | Codon, Amino Acid Change | Comments | Unique ID | A% | F% | Ref | CpG | S/M | Mut. type |
| New | 2418A>C | Y-16S | 1 | (95) | N | S | MM | |||
| New | 2436T>C | L-10P | 2 | (95) | N | S | MM | |||
| New | 2455C>A | C-4X | 3 | 50 | 50 | (96) | N | S | NM | |
| New | 2463delGinsTC | -1 | frameshift; stop codon 32 | 4 | 40 | 33 | (1) | N | S | FD/FI |
| New | 2487-8*insG | 8 | frameshift; stop codon 32 | 5 | (2) | N | S | FI | ||
| New | 2510-2512*delC | 16 | frameshift; stop codon 17 | 6 | 70 | (2) | N | S | FD | |
| New | 2527C>A | C21X | Loses BspW I site | 7 | (95) | N | S | NM | ||
| New | 2571-2580delCAGAA | 36-37, or 38-39 | frameshift; stop codon 70 | 8 | 57 | (95) | N | S | FD | |
| New | 2599-600*delC | 45 or 46 | frameshift; stop codon 81 | 9 | 46 | 49 | (3) | N | S | FD |
| 2601-2*delG | 46 | frameshift; stop codon 81 | 10 | 25/37 | (4) | N | S | FD | ||
| 2606insT | 48 | frameshift; stop codon 72 | 11 | 45 | 40/43 | (5) | N | S | FI | |
| 2610-31del22bp insA | 49-56 | Creates Acc I site | 12 | (6) | N | S | ID/I | |||
| 2627-29delAAT | 55 | Deletes N;Double mutation, also R57C | 13 | 60 | 56 | (6) | N | S | ID | |
| 2633C>T | R57C | Double mutation, also 55(-AAT)Loses Fau I site | 14 | (6) | Y | S | MM | |||
| 2638T>G | F58L | Type I or II? Budapest 6 | 15 | 45 | 55 | (95) | N | S | MM | |
| New | 2652A>G | Y63C | 16 | (2) | N | S | MM | |||
| New | 2652A>C | Y63S | Creates DdeI site | 17 | 42 | 36 | (1) | N | S | MM |
| New | 2661T>C | L66P | Loses AlwN I, BstN I, EcoR I sites and gains BcN I, Nci I, Apa I and Msp I sites | 18 | 55 | (95) | N | S | MM | |
| 2690-95delATTTTC | 76-77 | deletes I, F | 19 | 49 | 49/51 | (3) | N | S | ID | |
| New | 2702C>A | P80T | 20 | 63 | 54 | (7) | N | S | MM | |
| 2705-6delCT | 81 | frameshift; stop codon 95 | 21 | 68 | 59 | (6) | N | S | FD | |
| 2706delT | 81 | frameshift; stop codon 89 | 22 | 53 | 47 | (8) | N | S | FD | |
| New | 2765C>A | Q101K | 23 | 50 | 50 | (9) | N | S | MM | |
| 2770insT | 102 | frameshift; stop codon 107 | 24 | 43 | 43/51 | (4) | N | M (2) | FI | |
| 2770insT | 102 | frameshift; stop codon 107 | 25 | 43 | 41/50 | (4) | ||||
| New | 2777G>C | 1st nt. intron 2loses Mnl I, Rsa I, Csp6 I sites and gains Alu I site | 26 | 50 | 55 | (95) | N | S | SS | |
| New | 5306G>C | last nt. intron 2Loses Sfe I site and gains Mae II, SnaB I and BsaA I sites | 27 | (95) | N | S | SS | |||
| 5311-5320*del6bp | 106-108 | deletes F and K | 28 | 60 | 51 | (10) | N | M (3) | ID | |
| 5311-5320*del6bp | 106-108 | deletes F and K | 29 | (10) | ||||||
| 5311-5320*del6bp | 106-108 | deletes F and K | 30 | 52 | 64 | (11) | ||||
| 5352delT | 119 | frameshift; stop codon 126 | 31 | 60 | 63 | (12) | N | S | FD | |
| New | 5354C>T | H120Y | 32 | 62 | 69 | (7) | N | S | MM | |
| 5356-64*delCTT | 120-123 | deletes a F | 33 | 73 | 62 | (6) | N | M (2) | ID | |
| New | 5356-64*delCTT | " | deletes a F | 34 | 48 | 47 | (11) | |||
| New | 5373T>C | L126P | 35 | 52 | 54 | (9) | N | S | MM | |
| New | 5379G>A | C128Y | high MW complex | 36 | 60 | 65 | (13) | N | S | MM |
| 5381C>T | R129X | 37, 38 | <60 | <60 | (14) | Y | M (9) | NM | ||
| 5381C>T | " | 39-42 | <60 | <60 | (15) | |||||
| 5381C>T | " | 43 | (96) | |||||||
| 5381C>T | " | 44 | 50 | 50 | (11) | |||||
| New | 5381C>T | " | 45 | 49 | 50 | (1) | ||||
| New | 5390C>T | R132X | removes Taq I site | 46 | 50 | 50 | (9) | Y | M (2) | NM |
| 5390 C>T | " | 47 | 45 | 50 | (96) | |||||
| 5415T>A | L140X | 48 | 50 | 68 | (16) | N | S | NM | ||
| New | 5448-50*delC | 151-152 | frameshift; stop codon 251 | 49 | 52 | 56 | (17) | N | S | FD |
| New | 5492T>C | Y166H | Creates Rsa I, Nla III sites | 50 | 60 | 59 | (97) | N | S | MM |
| Change | 5493A>G | Y166C | Previously known as Whitechapel | 51 | 55 | 50 | (18) | N | M (3) | MM |
| 5493A>G | " | 52 | 49 | 46 | (6) | |||||
| New | 5493A>G | " | 53 | 52 | 55 | (95) | ||||
| New | 5501insA | 169 | frameshift; stop codon 192 | 54 | 42 | 40 | (1) | N | S | FI |
| 5524G>A | 176 | exon 3A donor splice site, transcript from mutant allele lacks exon 3A | 55 | 60 | 58 | (19) | N | S | SS | |
| New | 6431A>G | 5’ intron-exon splice site exon 3B. Alternative splice site creates stop codon 193 | 56 | 40 | 47 | (98) | N | S | SS | |
| New | 6457-59delATC | 186 | deletes an Ile,loses PfIM I, BsiY I sites | 57 | 50 | 56 | (2) | N | S | ID |
| 6462C>A | N187K | 58 | 121 | (20) | N | S | MM | |||
| New | 6484G>T | E195X | also GGC to GCC (G196A) | 59 | 56/59 | (2) | N | S | NM | |
| 6490C>T | R197X | 60 | 52 | 67 | (16) | Y | M (2) | NM | ||
| New | 6490C>T | " | 61 | (21) | ||||||
| 6523-24*insA | 208 | frameshift, stop codon 209 | 62 | 52 | 51/51 | (5) | N | S | FI | |
| New | 7384T>C | W225R | 63 | (97) | N | S | MM | |||
| 7392-94*insA | 228 | frameshift, stop codon 232 | 64 | <60 | <60 | (22) | N | S | FI | |
| 7428-29delCT | 239-240 | frameshift, stop codon 242 | 65 | 43 | 49/30 | (5) | N | S | FD | |
| 7445delA | 245 | frameshift, stop codon 251 | 66 | 54 | 53 | (23) | N | S | FD | |
| 7443-46*delAG | 244-245 | frameshift, stop codon 264 | 67 | 54 | 53 | (23) | N | S | FD | |
| New | 7520T>C | L270P | creates Dde I site | 68 | 50 | 47 | (9) | N | S | MM |
| New | 7522G>T | Q271X | loses Tth111 II site | 69 | (99) | N | S | NM | ||
| 7580-83*delAG | 290-291 | frameshift, stop codon 309 | 70 | <60 | <60 | (22) | N | S | FD | |
| 7596delA or delG | 295 | frameshift, stop codon 314Valine sequence variation site | 71 | 56 | 65 | (6) | N | S | FD | |
| 7634-39*del4bp | 308-309 | frameshift, stop codon 313 | 72 | <60 | <60 | (22) | N | S | FD | |
| 7644-49*delGGA | 311-313 | deletes a E | 73 | 47 | 57 | (6) | N | S | ID | |
| 7756T>C | S349P | 74 | 60 | 40 | (24) | N | S | MM | ||
| New | 7671delG | 320 | frameshift, stop codon 331 | 75 | 50 | 45 | (1) | N | S | FD |
| New | 7768-69delG | 353 | affects exon/intron 4 splice site and abolishes Bst NI site | 76 | 46 | 46 | (1) | N | S | SS |
| Change | 9788G>A | creates cryptic splice site and high MW complex, pleiotropic effect? | 77 | <60 | <60 | (13) | Y | M (3) | SS | |
| 9788G>A | creates cryptic splice site, removes Hpa II site | 78 | 52 | 59 | (6) | |||||
| New | 9788G>A | creates cryptic splice site | 79 | 64 | 58 | (17) | ||||
| 9819C>T | R359X | 80 | (6) | Y | M (2) | NM | ||||
| 9819 C>T | " | 81 | 44 | 43 | (97) | |||||
| 9852-53*delA | 370 | frameshift, stop codon 375 | 82 | 50 | 61/69 | (5) | N | S | FD | |
| New | 9858-60*delT | 372 | frameshift, stop codon 375 | 83 | 55 | 51 | (2) | N | S | FD |
| New | 9867G>T | first nt. intron 5 | 84 | 50 | (2) | N | S | SS | ||
| New | 13258T>G | S380R | 85 | 51 | 43 | (97) | N | S | MM | |
| New | 13277insACCG | 387 | frameshift, stop codon 433 | 86 | 52 | 50 | (25) | N | S | FI |
| 13278C>T | A387V | loses BspW I and NspB II sites | 87 | 60 | 50 | (26) | N | S | MM | |
| 13342insA | 408 | frameshift, stop codon 432 | 88 | <60 | <60 | (14) | N | M (2) | FI | |
| 13342insA | " | frameshift, stop codon 432 | 89 | (13) | ||||||
| 13347-51*delT | 410-411 | frameshift, stop codon 412 | 90 | (6) | N | S | FD | |||
| New | 13354delAA | 412 | frameshift, stop codon 431 | 91 | 46 | 58 | (25) | N | S | FD |
| 13354delAAGAG | 412-414 | frameshift, stop codon 430 | 92 | 43 | 30 | (11) | N | S | FD | |
| 13379insA | 421 | frameshift, stop codon 432 | 93 | 73 | (6) | N | S | FI | ||
| New | 13380T>C | I421T | abolishes Mbo II site | 94 | 46 | 45 | (1) | N | M (2) | MM |
| New | 13380T>C | " | abolishes Mbo II site | 95 | 68 | 68 | (17) | |||
| New | 13388G>C | G424R | gains Fsp I, Hha I and HinP I sites | 96 | 51 | 48 | (27) | N | S | MM |
| 13387-89*insG | 423-424 | frameshift, stop codon 432 | 97 | 75 | 52 | (8) | N | S | FI | |
| Change | 13397-405del9bp | 427-429 | deletes A, N, P also R47H. Forms high MW complex, pleiotropic effect? | 98 | (13) | N | S | ID | ||
| 13398C>A | A427D | gains Acc I site | 99 | 64 | 48 | (11) | N | S | MM | |
| New | 13407G>A | C430F | 100 | 60 | 55 | (1) | N | S | MM | |
| New | 13412-4insA | 432 | frameshift, adds extra 18 residues to C terminus | 101 | 50 | 50 | (98) | N | S | FI |
In this and other tables:
A%, antigen assay result as % of normal control
F%, functional assay result.
Two results indicate two types of assay were used.Nucleotide sequence numbering (28) makes no allowance for the highly polymorphic short tandem repeat region of intron 5 (29).
* Because of the repeat nature of the sequence the nucleotides affected cannot be unequivocally assigned.CpG mutation (CpG)- N no;Y yes.Single/multiple reports (S/M) - S single; M multiple, number of reports in brackets.Mutation type (Mut. type)- NM nonsense mutation; MM missense mutation; SS splice site mutation; FI frameshift insertion; FD frameshift deletion; ID inframe deletion; FD/FI deletion and insertion resulting in frameshift; ID/I deletion and insertion remaining in frame.
|
Deletion |
Unique Identifier |
Ref |
|
|
Deletes 2761 bp, including exon 5, from 9060-9104 to 11822-11865* |
102 |
(28) |
|
|
Deletes 5’-end; breakpoint 480bp upstream from 5’ boundary of exon 3a, 5’ to 5366 |
103 |
(30) |
|
|
New |
Deletes 105bp between 7432 and 7536, resulting in deletion of 35 amino acids, K241 to K275. Forms high molecular weigh complex, pleiotropic effect? |
104 |
(13) |
|
Deletes 5’ end, exons 1,2 |
105 |
(31) |
|
|
Removes 3’ end of gene, including exon 6 |
106 |
(31) |
|
|
Removes 3’ end of gene, including exon 6 |
107 |
(31) |
|
|
Uncharacterised |
108 |
(98) |
|
|
Whole gene deletion |
109 |
(32) |
|
|
Whole gene deletion |
110 |
(32) |
|
|
Whole gene deletion |
111 |
(33, 34) |
|
|
New |
Whole gene deletion |
112 |
(97) |
|
New |
Whole gene deletion |
113 |
(97) |
*Precise deletion points ambiguous
|
Nucleotide position, Mutation |
Codon, Amino acid change |
Variant; Unique Identifier |
A% |
F% |
Comments additional data in brackets |
Ref |
CpG |
M/S |
Mut. Type |
|
|
2484T>A |
I7N |
Rouen III; 1 |
Additional carbohydrate |
(57) |
N |
M (3) |
MM |
|||
|
2484T>A |
" |
2 unnamed families; 2, 3 |
Also V-3E |
(98) |
||||||
|
2534C>T |
R24C |
Rouen IV; 4 |
105 |
56 |
loses BsiY I site |
(58) |
Y |
S |
MM |
|
|
2586C>T |
P41L |
Basel; 5 |
104 |
60 |
loses Bcn I, Nci I, Msp I and Hpa II sites and gains BstN I, EcoR II and Gsu I sites |
(59) |
Y |
M (18) |
MM |
|
|
2586C>T |
" |
Clichy I; 6 |
110 |
68 |
" |
(41) |
||||
|
2586C>T |
" |
Clichy 2; 7 |
" |
(41) |
||||||
|
2586C>T |
" |
Franconville; 8 |
" |
(41) |
||||||
|
2586C>T |
" |
2 unnamed families; 9, 10 |
" |
(101) |
||||||
|
2586C>T |
" |
4 unnamed families; 11-13 |
" |
(38) |
||||||
|
2586C>T |
" |
Dublin 2; 14 |
" |
(60) |
||||||
|
2586C>T |
" |
unnamed; 15 |
" |
(98) |
||||||
|
2586C>T |
" |
unnamed; 16 |
" (Also V-3E) |
(98) |
||||||
|
New |
2586C>T |
" |
unnamed; 17 |
100 |
47 |
" |
(61) |
|||
|
New |
2586C>T |
" |
unnamed; 18 |
90 |
52 |
" |
(98) |
|||
|
New |
2586C>T |
" |
2 unnamed families; 19. 20 |
100 |
70-75 |
" |
(96) |
|||
|
New |
2586C>T |
" |
unnamed; 21 |
115 |
69 |
" |
(97) |
|||
|
2603C>T |
R47C |
Toyama; 22 |
100 |
26 |
(Homozygous) gains Age I site |
(62) |
Y |
M (19) |
MM |
|
|
2603C>T |
" |
Tours; 23 |
" |
(63) |
||||||
|
2603C>T |
" |
Alger; 24 |
Homozygous " |
(64) |
||||||
|
2603C>T |
" |
Paris 1; 25 |
127 |
54 |
" |
(41) |
||||
|
2603C>T |
" |
Paris 2; 26 |
100 |
60 |
" |
(41) |
||||
|
2603C>T |
" |
3 unnamed families; 27-29 |
" |
(101) |
||||||
|
2603C>T |
" |
Padua 2; 30 |
100 |
60 |
" |
(65) |
||||
|
2603C>T |
" |
unnamed; 31 |
" |
(38) |
||||||
|
2603C>T |
" |
Barcelona 2; 32 |
100 |
60 |
" |
(66) |
||||
|
2603C>T |
" |
Amiens; 33 |
" |
(67) |
||||||
|
2603C>T |
" |
Kumamoto; 34 |
100 |
28 |
(Homozygous) " |
(68) |
||||
|
2603C>T |
" |
unnamed; 35 |
" |
(98) |
||||||
|
2603C>T |
" |
2 unnamed families; 36, 37 |
90 |
60 |
" |
(95) |
||||
|
New |
2603C>T |
" |
Omura; 38 |
109 |
16 |
(Homozygous) " |
(100) |
|||
|
New |
2603C>T |
" |
Sasebo; 39 |
140 |
70 |
" |
(100) |
|||
|
New |
2603C>T |
" |
unnamed; 40 |
106 |
45 |
" |
(97) |
|||
|
2603C>A |
R47S |
Rouen II; 41 |
102 |
64 |
loses Cfr 10 I site |
(69) |
N |
S |
MM |
|
|
2604G>A |
R47H |
Rouen I; 42 |
111 |
55 |
gains Nsp I/NspC I, Nsp7524 I and Nla III sites |
(70) |
Y |
M (15) |
MM |
|
|
2604G>A |
" |
Padua I; 43 |
95 |
73 |
" |
(71) |
||||
|
2604G>A |
" |
Bligny; 44 |
100 |
60 |
" |
(72) |
||||
|
2604G>A |
" |
2 unnamed families; 45, 46 |
" |
(38) |
||||||
|
2604G>A |
" |
3 unnamed families; 47-49 |
" |
(101) |
||||||
|
2604G>A |
" |
unnamed; 50 |
" |
(25) |
||||||
|
2604G>A |
" |
3 unnamed families; 51-53 |
" |
(98) |
||||||
|
2604G>A |
" |
unnamed; 54 |
115 |
75 |
" |
(95) |
||||
|
2604G>A |
" |
Budapest 2; 55 |
" |
(95) |
||||||
|
New |
2604G>A |
" |
unnamed; 56 |
" |
(95) |
|||||
|
2759C>T |
L99F |
Budapest 3; 57 |
77 |
(Homozygous) loses Mnl I and Gsu I sites |
(73) |
N |
M (5) |
MM |
||
|
2759C>T |
" |
Budapest 7; 58 |
70 |
70 |
" |
(95) |
||||
|
2759C>T |
" |
unnamed; 59 |
54-75 |
19-54 |
(Homozygous) " |
(95) |
||||
|
2759C>T |
" |
unnamed; 60 |
71 |
55 |
" |
(95) |
||||
|
New |
2759C>T |
" |
unnamed; 61 |
55 |
56 |
" |
(96) |
|||
|
New |
2759C>G |
L99V |
Southport; 62 |
56 |
68 |
Variant detected by CIE loses Mnl I and Gsu I sites, gains Tth111 I site |
(61) |
N |
S |
MM |
|
New |
5342T>C |
S116P |
Nagasaki; 63 |
110 |
55 |
(74) |
N |
S |
MM |
|
|
New |
5349A>C |
Q118P |
Vienna; 64 |
104 |
52 |
loses Bcl I and BstY I sites and gains Alw I site - last of little diagnostic value as another site only 5bp away |
(61) |
N |
S |
MM |
|
5382G>A |
R129Q |
Geneva; 65 |
100 |
50 |
loses Hinf I and Ple I sites |
(75) |
Y |
M (4) |
MM |
|
|
5382G>A |
" |
unnamed; 66 |
" |
(101) |
||||||
|
5382G>A |
" |
unnamed; 67 |
" |
(98) |
||||||
|
5382G>A |
" |
unnamed; 68 |
169 |
75 |
" |
(95) |
||||
|
7420G>A |
E237K |
Truro; 69 |
gains Mbo II and Xmn I |
(102) |
N |
S |
MM |
Entries in bold and italics were the first report of this mutation
CpG - N no;Y yes.
Single/multiple reports (S/M) - S single; M multiple, number of reports in brackets.
Mut type Mutation type - MM missense mutation
|
Nucleotide position, Mutation |
Codon, Amino acid change |
Variant; Unique Identifier |
A% |
F% |
Comments |
Ref |
CpG |
M/S |
Mut. Type |
|
|
13323T>G |
F402C |
Rosny; 1 |
70 |
52 |
(76, 77) |
N |
S |
MM |
||
|
13323T>C |
F402S |
Torino; 2 |
69 |
46 |
gains BsaJ I and Sty I sites |
(76, 77) |
N |
S |
MM |
|
|
13324C>A |
F402L |
Maisons Laffitte; 3 |
73 |
56 |
gains Dra I and Mse I sites |
(77) |
N |
M (2) |
MM |
|
|
13324C>A |
" |
unnamed; 4 |
73 |
62/41 |
" |
(78) |
||||
|
13328G>A |
A404T |
Oslo; 5 |
48 |
50 |
loses Hae III and Hae I sites - of little diagnostic value as another site only 5bp away |
(77, 79) |
N |
M (3) |
MM |
|
|
13328G>A |
" |
Paris 3; 6 |
72 |
48 |
" |
(77) |
||||
|
13328G>A |
" |
unnamed; 7 |
100 |
50/65 |
" |
(11) |
||||
|
13333C>G |
N405K |
La Rochelle; 8 |
73 |
55 |
(Also V-3E) gains Mnl I site |
(77) |
N |
M (2) |
MM |
|
|
13333C>G |
" |
1 family; 9 |
32/57 |
" |
(98) |
|||||
|
New |
13334A>G |
R406G |
unnamed; 10 |
78 |
32 |
gains Sau 96 I site, loses Hae I and Stu I sites |
(96) |
N |
S |
MM |
|
13335G>T |
R406M |
Kyoto; 11 |
Described as type I deficiency loses Stu I site and gains Nsp I/NspC I and Nsp7524 sites |
(80) |
N |
S |
MM |
|||
|
13337C>A |
P407T |
Budapest 5; 12 |
100 |
70 |
(Also Protein C deficiency) loses Stu I, Hae I and Hae III sites - last 2 of little diagnostic value as another site only 5bp away |
(77) |
N |
M (2) |
MM |
|
|
New |
13337C>A |
" |
unnamed; 13 |
86 |
72 |
" |
(96) |
|||
|
13338C>T |
P407L |
Utah; 14 |
50 |
50 |
loses Stu I, Hae I and Hae III sites - last 2 of little diagnostic value as another site only 5bp away |
(81) |
N |
M (2) |
MM |
|
|
New |
13338C>T |
" |
unnamed; 15 |
" |
(95) |
|||||
|
New |
13392G>C |
R425T |
unnamed; 16 |
37 |
42/45 |
gains Bsp 1286 I site |
(96) |
N |
M (2) |
MM |
|
New |
13392G>C |
" |
unnamed; 17 |
50-56 |
50-59 |
" |
(103) |
|||
|
13404C>T |
P429L |
Budapest; 18 |
75 |
20 |
Homozygous, but 2 plasma forms |
(82) |
N |
S |
MM |
|
Entries in bold and italics were the first report of this mutation
CpG mutation (CpG)- N no;Y yes.
Single/multiple reports (S/M) - S single; M multiple, number of reports in brackets.
Mutation type (Mut. type)- MM missense mutation
|
Nucleotide position, Mutation |
Codon, Amino acid change |
Variant; Unique Identifier |
A% |
F% |
Comments additional data in brackets |
Ref |
CpG |
S/M |
Mut. type |
|
|
6460A>G |
N187D |
Rouen VI; 1 |
160 |
51/65 |
Increased heparin affinity Forms polymers |
(35) |
N |
M (3) |
MM |
|
|
6460A>G |
" |
unnamed; 2 |
98 |
55 |
(20) |
|||||
|
6460A>G |
" |
unnamed;3 |
77 |
62 |
(96) |
|||||
|
New |
7464G>A |
M251I |
unnamed;4 |
73 |
52 |
(7) |
N |
S |
MM |
|
|
7562T>A |
I284N |
Haslar;5 |
69 |
64/82 |
(98) |
N |
S |
MM |
||
|
New |
7615G>A |
E302K |
unnamed; 6 |
88 |
70 |
loses Mnl I site |
(98) |
N |
S |
MM |
|
13262G>A |
A382T |
Hamilton; 7 |
100 |
50 |
(Transformed into substrate) loses Bbv I site and Fnv4H I site |
(36, 37) |
Y |
M (4) |
MM |
|
|
13262G>A |
" |
unnamed; 8 |
" |
(38) |
||||||
|
13262G>A |
" |
Glasgow II; 9 |
100 |
56 |
(Transformed into substrate) |
(39) |
||||
|
13262G>A |
" |
unnamed; 10 |
108 |
28/71 |
" |
(98) |
||||
|
13268G>C |
A384P |
Charleville; 11 |
100 |
60 |
(Transformed into substrate) loses Bbv I site, Fnv4H I site, BspW I site |
(40, 41) |
N |
M (6) |
MM |
|
|
13268G>C |
Cambridge I; 12 |
95 |
54 |
" |
(42) |
|||||
|
13268G>C |
" |
Vicenza; 13 |
105 |
62 |
(Transformed into substrate) " |
(40) |
||||
|
13268G>C |
" |
unnamed; 14 |
" |
(38) |
||||||
|
13268G>C |
" |
Sudbury; 15 |
119 |
56 |
" |
(43) |
||||
|
New |
13268G>C |
" |
unnamed; 16 |
100 |
60 |
" |
(95) |
|||
|
13268G>T |
A384S |
Cambridge II; 17 |
103 |
75 |
loses Bbv I site, Fnv4H I site, BspW I site and Pvu II site |
(44) |
N |
M (20) |
MM |
|
|
13268G>T |
" |
unnamed; 18 |
" |
(98) |
||||||
|
13268G>T |
" |
unnamed; 19 |
113 |
59 |
" |
(98) |
||||
|
13268G>T |
" |
unnamed; 20 |
108 |
" |
(98) |
|||||
|
13268G>T |
" |
13 unnamed cases; 21-33 |
94 |
95/76 |
" (all but 3 asymptomatic) |
(98) |
||||
|
13268G>T |
" |
unnamed; 34 |
100 |
74 |
" |
(95) |
||||
|
New |
13268G>T |
" |
unnamed; 35 |
90 |
75 |
" |
(96) |
|||
|
New |
13268G>T |
" |
unnamed; 36 |
90 |
74 |
" |
(98) |
|||
|
13293G>A |
G392D |
Stockholm; 37 |
loses Eae I site,Gdi II site, Hae III site and BceF I site |
(45) |
Y |
S |
MM |
|||
|
13295C>T |
R393C |
Northwick Park; 38 |
162 |
65 |
complex with albumin loses Eae I site,Gdi II site, Hae III site and BceF I site |
(46, 47) |
Y |
M (3) |
MM |
|
|
13295C>T |
" |
Milano I; 39 |
" |
(48) |
||||||
|
13295C>T |
" |
Frankfurt I; 40 |
100 |
60 |
" |
(49,50) |
||||
|
13296G>A |
R393H |
Glasgow; 41 |
87 |
43 |
(Increased heparin affinity) loses Gdi II site and BceF I sites and gains Hae I and Msc I sites |
(47) |
Y |
M (7) |
MM |
|
|
13296G>A |
" |
Sheffield; 42 |
75 |
54 |
" |
(51) |
||||
|
13296G>A |
" |
Chicago; 43 |
96 |
37 |
(Increased heparin affinity) " |
(52) |
||||
|
13296G>A |
" |
Avranches; 44 |
100 |
59 |
" |
(41) |
||||
|
13296G>A |
" |
unnamed; 45 |
" |
(98) |
||||||
|
New |
13296G>A |
" |
unnamed; 46 |
93 |
56 |
" |
(96) |
|||
|
New |
13296G>A |
" |
Kumamoto II; 47 |
92 |
61 |
(Increased heparin affinity) " |
(100) |
|||
|
13296G>C |
R393P |
Pescara; 48 |
100 |
62 |
Increased heparin affinity loses Eae I site, Gdi II site, BceF I site and gains Sau96 I site |
(53, 54) |
N |
S |
MM |
|
|
13299C>T |
S394L |
Denver,49 |
92 |
54 |
increased clearance ? |
(55) |
Y |
M (7) |
MM |
|
|
13299C>T |
" |
Milano 2; 50 |
(56) |
|||||||
|
13299C>T |
" |
unnamed; 51 |
123 |
68 |
(11) |
|||||
|
New |
13299C>T |
" |
4 unnamed cases; 52-55 |
100 |
66-71 |
(7) |
Entries in bold were the first report of this mutation.
CpG mutation (CpG)- N no;Y yes.
Single/multiple reports (S/M)) - S single; M multiple, number of reports in brackets.
Mutation type (Mut.Type) - MM missense mutation
|
Nucleotide Position, Sequence |
Location |
Detection |
Comment |
Ref |
|
|
(-276), 76bp dimorphism |
5’ untranslated region |
* |
Slow 0.25, Fast 0.75 |
(83) |
|
|
(-159), T/C |
" |
DGGE |
T 0.96, C 0.04 *** |
(95) |
|
|
67G>A |
" |
Very low; found in deficient family and not in 78 normal alleles |
(6) |
||
|
2457T/A |
Exon 2; -3V/E |
sequencing |
4 families |
(60) |
|
|
" |
" |
3 families, one also N405K |
(95) |
||
|
" |
" |
4 families, 2 also I7N, one also P41L |
(98) |
||
|
" |
" |
1 family |
(96) |
||
|
" |
" |
1 family with type I deficiency |
(11) |
||
|
New |
2523T/C |
Exon 2; 20M/T |
sequencing |
Previously Whitechapel |
(20) |
|
New |
5469A/G |
Exon 3a; 158Y/C |
sequencing |
4 families, 3 of Danish origin |
(7) |
|
7596G/A |
Exon 4; 295V |
sequencing |
GTG 0.43**, GTA 0.57 |
(82) |
|
|
7626G/A |
Exon 4; 305Q |
Pst I |
+ 0.5,-0.5 |
(33) |
|
|
7987T/C |
Intron 4 |
Nhe I |
+0.64,-0.36 |
(84) |
|
|
9820G/A |
Exon 5; R/Q |
sequencing |
1 family |
(102) |
|
|
8136-8153 (ATT)n |
Intron 4; Alu 5 tail |
sequencing |
n = 6,8 |
(29) |
|
|
9289-9333 (ATT)n |
Intron 4; Alu 8 tail |
sequencing |
Highly polymorphic n = 5-18 |
(95) |
|
|
9893C/G |
Intron 5 |
Dde I |
= +0.83,-0.17 |
(85) |
* can be detected by visualisation of amplified DNA fragments spanning the polymorphism (8, 86)
** based upon sequencing 20 alleles
*** based on analyzing 156 alleles by denaturing gradient gel electrophoresis (DGGE)
+ presence of restriction enzyme cutting site, - absence of restriction enzyme cutting site
|
Distinct mutations |
Total database entries |
|
|
Missense mutation |
60 (35) |
171 (143) |
|
Nonsense mutation |
8 (0) |
19 (0) |
|
Inframe deletion |
7 (0) |
10 (0) |
|
Frameshift deletion |
20 (0) |
20 (0) |
|
Frameshift insertion |
11 (0) |
13 (0) |
|
Splice site mutation |
7 (0) |
9 (0) |
|
Major gene deletion |
12 (0) |
12 (0) |
|
Frameshift deletion/Frameshift insertion |
1 (0) |
1 (0) |
|
Inframe deletion/insertion |
1 (0) |
1 (0) |
|
Totals |
127 (35) |
256 (143) |
"Distinct mutations" do not include those repeated mutations that may have arisen independently
The entries are of total numbers in the database, while those in brackets refer to type II deficiency mutations.
|
Distinct |
Total |
||
|
I |
|||
|
Point mutations |
80 |
101 |
|
|
Partial/whole deletions |
12 |
12 |
|
|
II |
|||
|
RS |
12 |
55 |
|
|
HBS |
12 |
70 |
|
|
PE |
11 |
18 |
|
|
Totals |
127 |
256 |
RS : Reactive site mutations
HBS : Heparin binding site mutations
PE : Mutations with pleiotropic effects
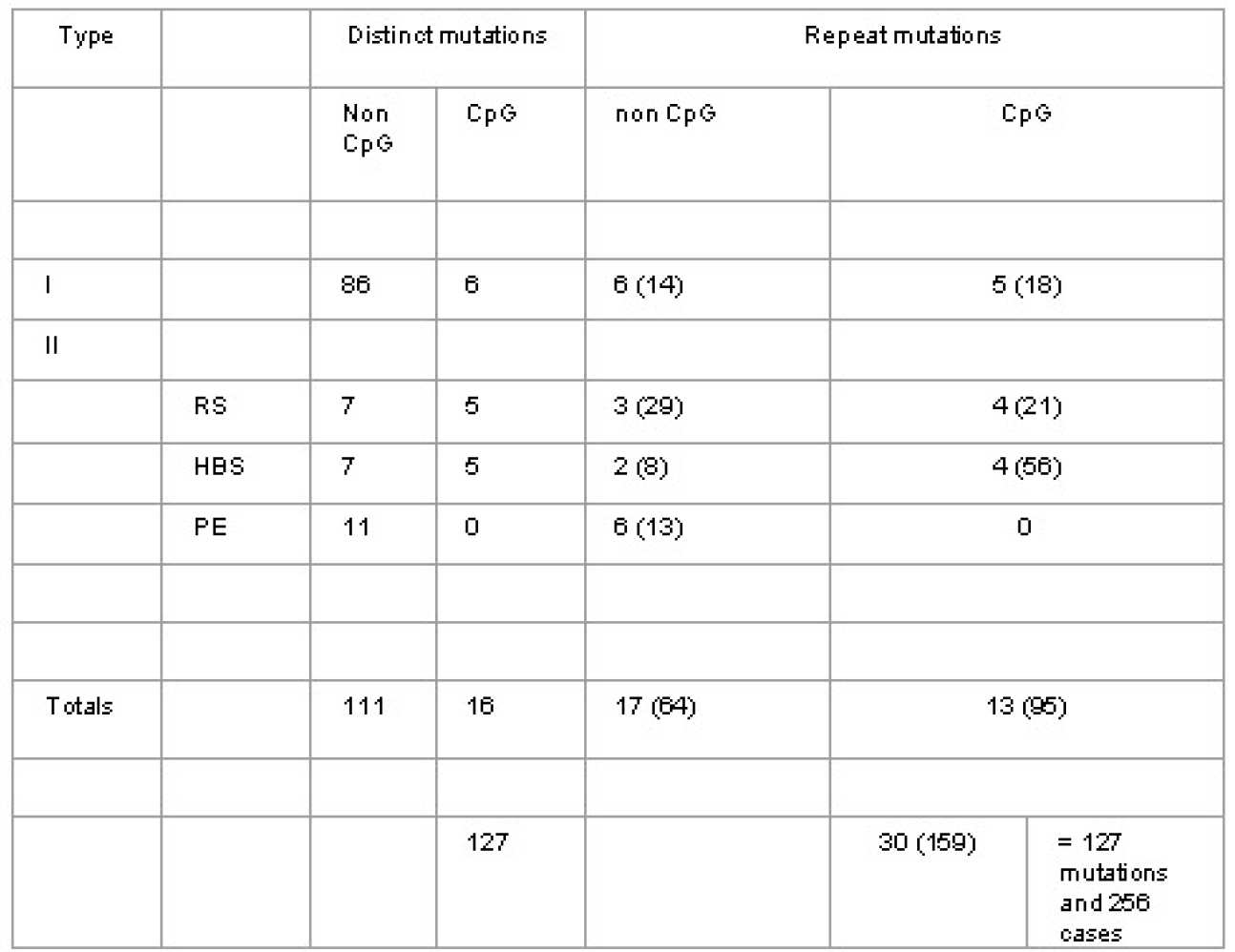
RS : Reactive site mutations
HBS : Heparin binding site mutations
PE : Mutations with pleiotropic effects " Distinct mutations" do not include those repeated mutations which may have arisen independantly Type I includes point mutations, partial and whole gene deletions Numbers in brackets include total number of entries in the database for each category CpG mutations are those involving CG to TG or CA substitution mutations.
1. van Boven HH, Olds RJ, Thein SL, Reitsma PH, Lane DA, Briet E, Vandenbroucke JP, Rosendaal FR. Hereditary antithrombin deficiency: heterogeneity of the molecular basis and mortality in Dutch families. Blood 1994; 84: 4209-4213.
2. Csurgay E, Ireland H, Thompson E, Conard J, Mannucci PM, Andrews V, Worsley A, Sas G, Lane DA. Novel mutations in seven families with type I antithrombin deficiency. Thrombosis and Haemostasis 1995; 73: 935.
3. Daly M, Perry DJ, Bruce DB, Harper PL, Tait RC, Walker ID, Mayne EE, Daly HM, Brown K, Carrell RW. Type I antithrombin deficiency: 5 novel mutations associated with thrombosis. Blood Coagulation and Fibrinolysis 1996.
4. Daly ME, Perry DJ, Harper PL, Carrell RW. The molecular basis of quantitative antithrombin deficiency in 8 unrelated families. British Journal of Haematology 1992; 80: 15.
5. Daly M, Perry DJ, Harper PL, Daly HM, Roques AWW, Carrell RW. Insertions/deletions in the antithrombin gene: 3 mutations associated with non-expression. Thrombosis and Haemostasis 1992; 67: 521-525.
6. Chowdhury V, Olds RJ, Lane DA, Conard J, Pabinger I, Ryan K, Bauer K, Bhavani M, Abildgaard U, Finazzi G, Castaman G, Mannucci PM, Thein SL. Identification of nine novel mutations in type I antithrombin deficiency by heteroduplex screening. British Journal of Haematology 1993; 84: 656-661.
7. Millar DS, Wacey AI, Ribando J, Melissari E, Laursen B, Woods P, Kakkar VV, Cooper DN. Three novel missense mutations in the antithrombin III (AT3) gene causing recurrent venous thrombosis. Human Genetics 1995; 94: 509-512.
8. Olds RJ, Lane DA, Ireland H, Leone G, De Stefano V, Wiesel ML, Cazenave JP, Thein SL. Novel point mutations leading to type I antithrombin deficiency and thrombosis. British Journal of Haematology 1991; 78: 408-413.
9. Perry DJ. Ectopic transcript analysis in human antithrombin deficiency. Blood Coagulation and Fibrinolysis 1995; 6: 531-536.
10. Olds RJ, Lane DA, Beresford CH, Abildgaard U, Hughes PM, Thein SL. A recurrent deletion in the antithrombin gene, AT106-108(-6bp), identified by DNA heteroduplex detection. Genomics 1993; 16: 298-299.
11. Millar DS, Lopez A, White D, Abraham G, Laursen B, Holding S, Reverter JC, Reynand J, Martinowitz U, Hayes JPLA, Kakkar VV, Cooper DN. Screening for mutations in the antithrombin III (AT3) gene causing recurrent venous thrombosis by single strand conformation polymorphism analysis. Human Mutation 1993; 2: 324-326.
12. Olds RJ, Lane DA, Finazzi G, Barbui T, Thein SL. A frameshift mutation leading to type 1 antithrombin deficiency and thrombosis. Blood 1990; 76: 2182-2186.
13. Emmerich J, Vidaud D, Alhenc-Gelas M, Chadeuf G, Gouault-Heilmann M, Aillaud MF, Aiach M. Three novel mutations of antithrombin inducing high molecular mass compounds. Arteriosclerosis and Thrombosis 1994; 14: 1958-1965.
14. Gandrille S, Vidaud D, Emmerich J, Clauser E, Sie P, Fiessinger JN, Alhenc-Gelas M, Priollet P, Aiach M. Molecular basis for hereditary antithrombin III quantitative deficiencies: a stop codon in exon IIIa and a frameshift in exon V1. British Journal of Haematology 1991; 78: 414-420.
15. Olds RJ, Lane DA, Ireland H, Finazzi G, Barbui T, Abildgaard U, Girolami A, Thein SL. A common point mutation producing type Ia antithrombin III deficiency: AT129 CGA to TGA (Arg to Stop). Thrombosis Research 1991; 64: 621-625.
16. Tomonori A, Iwahana H, Yoshimoto K, Shigekiyo T, Saito S, Itakura M. Two new nonsense mutations in type Ia antithrombin III deficiency at Leu140 and Arg197. Thrombosis and Haemostasis 1992; 68: 455-459.
17. Jochmans K, Lissens W, Yin T, Michiels JJ, van der Luit L, Peerlinc- K, Vervoot R, De Wale M, Liebaers I. Molecular basis for type I antithrombin deficiency: identification of two novel mutations and evidence for a de novo splice mutation. Blood 1994; 84: 3742-3748.
18. Perry DJ, Daly ME, Colvin BT, Brown K, Carrell RW. Two antithrombin mutations in a compound heterozygote: Met20Thr and Tyr166Cys. American Journal of Hematology 1995; 50: 215-216.
19. Berg LP, Grundy CB, Thomas F, Millar DS, Green PJ, Slomski R, Reiss J, Kakkar VV, Cooper DN. De novo splice site mutation in the antithrombin III (AT3) gene causing recurrent venous thrombosis: demonstration of exon skipping by ectopic transcript analysis. Genomics 1992; 13: 1359-1361.
20. Perry DJ, Marshall C, Borg JY, Tait RC, Daly ME, Walker ID, Carrell RW. Two new antithrombin variants, Asn 187 Lys, indicate a functional role for asparagine 187. Blood Coagulation and Fibrinolysis 1995; 6: 51-54.
21. Michiels JJ, van der Luit L, van Vliet H, Jochmans K, Lissens W. Nonsense mutation in Arg 197 stop in a Dutch family with type I hereditary antithrombin deficiency causing thrombophilia. Thrombosis Research 1995; 78: 251-254.
22. Vidaud D, Emmerich J, Sirieix ME, Sie P, Alhenc-Gelas M, Aiach M. Molecular basis for antithrombin III type I deficiency: 3 novel mutations located in exon IV. Blood 1991; 78: 2305-2309.
23. Grundy CB, Thomas F, Millar DS, Krawczak M, Melissari E, Lindo V, Moffat E, Kakkar VV, Cooper DN. Recurrent deletion in the human antithrombin III gene. Blood 1991; 78: 1027-1032.
24. Grundy CB, Holding S, Millar DS, Kakkar VV, Cooper DN. A novel missense mutation in the antithrombin III gene (Ser 349 to Pro) causing recurrent venous thrombosis. Human Genetics 1992; 88: 707-708.
25. Emmerich J, Chadeuf G, Alhenc-Gelas M, Gouault-Heilmann M, Toulon P, Fiessinger JN, Aiach M. Molecular basis of antithrombin type I deficiency: the first large in-frame deletion and two novel mutations in exon 6. Thrombosis and Haemostasis 1994; 72: 534-539.
26. White D, Abraham G, Carter C, Kakkar VV, Cooper DN. A novel missense mutation in the antithrombin III gene (Ala 387 to Val) causing recurrent venous thrombosis. Human Genetics 1993; 90: 472-473.
27. Jochmans K, Lissens W, Vervoot R, Peeters S, De Wale M, Liebaers I. Antithrombin-Gly 424 Arg: a novel point mutation responsible for type I antithrombin deficiency and neonatal thrombosis. Blood 1994; 83: 146-151.
28. Olds RJ, Lane DA, Chowdury V, De Stefano V, Leone G, Thein SL. Complete nucleotide sequence of the antithrombin gene. Evidence for homologous recombination causing thrombophilia. Biochemistry 1993; 32: 4216-4224.
29. Olds RJ, Lane DA, Chowdury V, Sas G, Pabinger I, Auberger K, Thein SL. (ATT) trinucleotide repeats in the antithrombin gene and their use in determining the origin of repeated mutations. Human Mutation 1994; 4: 31-41.
30. Fernandez-Rachubinski RA, Rachubinski RA, Blajchman MA. Partial deletion of antithrombin III allele in a kindred with a type I deficiency. Blood 1992; 80: 1476-1485.
31. Olds RJ, Lane DA, Chowdury V, Samson D, DeStafano V, Leone G, Wiesel ML, Cazenave JP, Conard J, Thein SL. Major rearrangements within the antithrombin gene locus: an unusual cause for antithrombin deficiency. British Journal of Haematology 1992; 40.
32. Winter JH, Bennett B, Watt JL, Brown T, San Roman C, Schinzel A, King J, Cooke PJL. Confirmation of linkage between antithrombin III and Duffy blood group and assignment of AT3 to 1q22-25. Annals of Human Genetics 1982; 26: 29-34.
33. Prochownik EV, Antonarakis S, Bauer KA, Rosenberg RD, Fearon ER, Orkin SH. Molecular heterogeneity of inherited antithrombin III deficiency. New England Journal of Medicine 1983; 308: 1549-1552.
34. Bock SC, Prochownik EV. Molecular genetic survey of 16 kindreds with hereditary antithrombin III deficiency. Blood 1987; 70: 1273-1278.
35. Bruce D, Perry DJ, Borg J-Y, Carrell RW, Wardell MR. Thromboembolic disease due to thermolabile conformational changes of antithrombin Rouen VI (187 Asn to Asp). Journal of Clinical Investigation 1994; 94: 2265-2274.
36. Devraj-Kizu- R, Chui DHK, Prochownik EV, Carter CJ, Ofosu FA, Blajchman MA. Antithrombin III Hamilton: a gene with a point mutation (guanine to adenine) in codon 382 causing impaired serine protease reactivity. Blood 1988; 72: 1518-1523.
37. Austin RC, Rachubinski RA, Ofosu FA, Blajchman MA. Antithrombin-III-Hamilton, Ala 382 to Thr: an antithrombin -III variant that acts as a substrate but not an inhibitor of a-thrombin and Factor Xa. Blood 1991; 77: 2185-2189.
38. Perry DJ, Carrell RW. CpG dinucleotides are "hotspots" for mutation in the antithrombin III gene. Twelve variants identified using the polymerase chain reaction. Molecular Biology in Medicine 1989; 6: 239-243.
39. Ireland H, Lane DA, Thompson E, Walker ID, Blench I, Morris HR, Freyssinet JM, Grunebaun L, Olds R, Thein SL. Antithrombin Glasgow II: alanine to threonine mutation in the serpin P12 position, resulting in a substrate reaction with thrombin. British Journal of Haematology 1991; 79: 70-74.
40. Caso R, Lane DA, Thompson EA, Olds RJ, Thein SL, Panico M, Blench I, Morris H, Freyssinet JM, Aiach M, Rodeghiero F, Finazzi G. Antithrombin Vicenza, Ala 384 to Pro (GCA to CCA) mutation transforming the inhibitor into a substrate. British Journal of Haematology 1991; 77: 87-92.
41. Mohlo-Sabatier P, Aiach M, Gaillard I, Fiessinger JN, Fischer AM, Chadeuf G, Clauser E. Molecular characterization of antithrombin III (AT III) variants using polymerase chain reaction. Identification of the ATIII Charleville as an Ala384 Pro mutation. Journal of Clinical Investigation 1989; 84: 1236-1241.
42. Perry DJ, Harper PL, Fairham S, Daly M, Carrell RW. Antithrombin Cambridge, 384 Ala to Pro: A new variant identified using the polymerase chain reaction. Federation of European Biochemical Societies Letters 1989; 254: 174-176.
43. Penwarchuk WJ, Fernandez-Rachubinski F, Rachubinski RA, Blajchman MA. Antithrombin III Sudbury, an Ala 384 to Pro mutation with abnormal thrombin binding activity and thrombotic diathesis. Thrombosis Research. 1990; 59: 793-798.
44. Perry DJ, Daly M, Harper PL, Tait RC, Price J, Walker ID, Carrell RW. Antithrombin Cambridge II, 384 Ala to Ser. Further evidence of the role of the reactive centre loop in the inhibitory function of the serpin. Federation of European Biochemical Societies Letters 1991; 285: 248-250.
45. Blajchman MA, Fernandez-Rachubinsky F, Sheffield WP, Austin RC, Schulman S. Antithrombin III Stockholm: a codon 392 (Gly to Asp) mutation with normal heparin binding and impaired serine protease reactivity. Blood 1992; 79: 1428-1434.
46. Erdjument H, Lane DA, Ireland H, Panico M, DiMarzo V, Blench I, Morris HR. Formation of a covalent disulfide-linked antithrombin complex by an antithrombin variant, antithrombin Northwick Park. Journal of Biological Chemistry 1987; 262: 13381-13384.
47. Erdjument H, Lane DA, Panico M, diMarzo V, Morris HR. Single amino acid substitutions in the reactive site of antithrombin leading to thrombosis. Congenital substitution of arginine 393 to cysteine in antithrombin Northwick Park and to histidine in antithrombin Glasgow. Journal of Biological Chemistry 1988; 263: 5589-5593.
48. Erdjument H, Lane DA, Ireland H, DiMarzo V, Panico M, Morris HR, Tripodi A, Mannucci PM. Antithrombin Milano, single amino acid substitution at the reactive site, Arg 393 to Cys. Thombosis and Haemostasis 1988; 60: 471-475.
49. Ireland H, Lane DA, Thompson E, Olds R, Thein SL, Hach-Wunderle V, Scharrer I. Antithrombin Frankfurt I: arginine to cysteine substitution at the reactive site and formation of a variant antithrombin-albumin covalent complex. Thrombosis and Haemostasis 1991; 65: 913.
50. van Boven HH, Reitsma PH, Rosendaal FR, Bayston TA, Chowdhury V, Bauer K, Scharrer I, Conard J, Lane DA. Factor V Leiden (FV R506Q) in families with inherited antithrombin deficiency. Thromb Haemost 1996; 75: 417-421.
51. Lane DA, Erdjument H, Flynn A, DiMarzo V, Panico M, Morris H, Greaves M, Dolan G, Preston FE. Antithrombin Sheffield: amino acid substitution at the reactive site (Arg 393 to His) causing thrombosis. British Journal of Haematology 1989; 71: 91-96.
52. Erdjument H, Lane DA, Panico M, DiMarzo V, Morris HR, Bauer K, Rosenberg RD. Antithrombin Chicago, amino acid substitution of arginine 393 to histidine. Thrombosis Research 1989; 54: 613-619.
53. Lane DA, Erdjument H, Thompson E, Panico M, DiMarzo V, Morris HR, Leone G, De Stefano V, Thein SL. A novel amino acid substitution in the reactive site of a congenital variant antithrombin. Antithrombin Pescara, Arg 393 to Pro, caused by CGT to CCT mutation. Journal of Biological Chemistry 1989; 264: 10200-10204.
54. Owen MC, George PM, Lane DA, Boswell DR. P1 variant antithrombins Glasgow (393 Arg to His) and Pescara (393 Arg to Pro) have increased heparin affinity and are resistant to catalytic cleavage by elastase. Implications for the heparin activation mechanism. Federation of European Biochemical Societies Letters 1991; 280: 216-220.
55. Stephens AW, Thalley BS, Hirs CHW. Antithrombin Denver, a reactive site variant. Journal of Biological Chemistry 1987; 262: 1044-1048.
56. Olds RJ, Lane D, Caso R, Tripodi A, Mannucci PM, Thein SL. Antithrombin III Milano 2: a single base substitution in the thrombin binding domain detected with PCR and direct genomic sequencing. Nucleic Acids Research 1989; 17: 10511.
57. Brennan SO, Borg JY, George PM, Soria C, Soria J, Caen J, Carrell RW. New carbohydrate site in mutant antithrombin (7Ile-Asn) with decreased heparin affinity. Federation of European Biochemical Societies Letters 1988; 237: 118-122.
58. Borg JY, Brennan SO, Carrell RW, George P, Perry DJ, Shaw J. Antithrombin Rouen IV 24 Arg to Cys. The amino terminal contribution to heparin binding. Federation of European Biochemical Societies Letters 1990; 266: 163-166.
59. Chang JY, Tran TH. Antithrombin Basel. Identification of a Pro-Leu substitution in a hereditary abnormal antithrombin with impaired heparin cofactor activity. Journal of Biological Chemistry 1986; 261: 1174-1176.
60. Daly M, Bruce D, Perry DJ, Price J, Harper PL, O'Meara A, Carrell RW. Antithrombin Dublin (-3 Val to Glu): an N-terminal variant which has an aberrant signal peptide cleavage site. Federation of European Biochemical Societies Letters 1990; 273: 87-90.
61. Chowdhury V, Mille B, Olds RJ, Lane DA, Watton J, Barrowcliffe TW, Pabinger I, Woodcock BE, Thein SL. Antithrombins Southport (99 Leu to Val) and Vienna (118 Gln to Pro): two novel antithrombin variants with abnormal heparin binding. British Journal of Haematology 1995; 89: 602-609.
62. Koide T, Odani S, Takahashi K, Ono T, Sakuragawa N. Antithrombin III Toyama; replacement of Arginine 47 by Cysteine in hereditary abnormal antithrombin III that lacks heparin-binding ability. Proceedings of the National Academy of Science USA 1984; 81: 289-293.
63. Duchange N, Chasse JF, Cohen GN, Zakin MM. Identification of a mutation leading to cysteine replacement in a silent deficiency. Nucleic Acids Research 1986; 14: 2408.
64. Brunel F, Duchange N, Fischer AM, Cohen GN, Zakin MM. Antithrombin III Alger: a new case of Arg 47 - Cys mutation. American Journal of Haematology 1987; 25: 223-224.
65. Olds RJ, Lane DA, Caso R, Girolami A, Thein SL. Antithrombin III Padua II: a single base substitution in exon 2 detected using PCR and direct genomic sequencing. Nucleic Acids Research 1990; 18: 1926.
66. Owen MC, Shaw GJ, Grau E, Fontcuberta J, Carrell RW, Boswell DR. Molecular characterisation of antithrombin Barcelona 2: 47 arginine to cysteine. Thrombosis Research 1989; 55: 451-457.
67. Fernandez-Rachubinski F, Rachubinski R, Blajchman MA. Genetic characterisation of kindreds with antithrombin III (AT-III) deficiency using selected amplification of the gene. Blood 1990; 76: 506a.
68. Ueyama H, Murakami T, Nishiguchi S, Maeda S, Hashimoto Y, Okajima K, Shimada K, Araki S. Antithrombin Kumamoto. Identification of a point mutation and genotype analysis of the family. Thrombosis and Haemostasis 1990; 63: 231-234.
69. Borg JY, Owen MC, Soria C, Soria J, Caen J, Carrell RW. Arginine 47 is a prime heparin binding site in antithrombin. A new variant Rouen II, 47 Arg to Ser. Journal of Clinical Investigation 1988; 81: 1292-1296.
70. Owen MC, Borg JY, Soria C, Soria J, Caen J, Carrell RW. Heparin binding defect in new antithrombin III variant: Rouen, 47 Arg to His. Blood 1987; 69: 1275-1279.
71. Caso R, Lane DA, Thompson E, Zangouras D, Panico M, Morris H, Olds RJ, Thein SL, Girolami A. Antithrombin Padua I: impaired heparin binding caused by an Arg 47 to His (CGT to CAT) substitution. Thrombosis Research 1990; 58: 185-190.
72. Wolf M, Boyer-Neumann C, Mohlo-Sabatier P, Neumann C, Meyer D, Larrieu MJ. Familial variant of antithrombin III (AT III Bligny, 47 Arg to His) associated with protein C deficiency. Thrombosis and Haemostasis 1990; 63: 215-219.
73. Olds RJ, Lane DA, Boisclair M, Sas G, Bock SC, Thein SL. Antithrombin Budapest 3: an antithrombin variant with reduced heparin affinity resulting from the substitution L99F. Federation of European Societies Letters 1992; 300: 241-246.
74. Okajima K, Abe H, Maeda S, Motomura M, Tsujihata M, Nagataki S, Okabe H, Takatsuki K. Antithrombin III Nagasaki (Ser116-Pro): a heterozygous variant with defective heparin binding associated with thrombosis. Blood 1993; 81: 1300-1305.
75. Gandrille S, Aiach M, Lane DA, Vidaud D, Mohlo-Sabatier P, Caso R, de Moerloose P, Fiessinger JN, Clauser E. Important role of Arg 129 in heparin binding site of antithrombin III: identification of novel mutation Arg 129 to Gln. Journal of Biological Chemistry 1990; 265: 18997-19001.
76. Olds RJ, Thein SL, Ireland H, Lane DA, Boisclair M, Conard J, Horellou MH. Identification of 402 phenylalanine as a functionally important residue in antithrombin. Thrombosis and Haemostasis 1991; 65: 670.
77. Lane DA, Olds RJ, Conard J, Boisclair M, Bock SC, Hultin M, Abildgaard U, Ireland H, Thompson E, Sas G, Horellou MH, Tamponi G, Thein SL. Pleiotropic effects of antithrombin strand 1C substitution mutations. Journal of Clinical Investigation 1992; 90: 2422-2433.
78. Emmerich J, Chadeuf G, Coetzee MJ, Alhenc-Gelas M, Friessinger J-N, Aiach M. A phenylalanine 402 to leucine mutation is responsible for a stable inactive conformation of antithrombin. Thrombosis Research 1994; 76: 307-315.
79. Bock SC, Silberman JA, Wikoff W, Abildgaard U, Hultin MB. Identification of a threonine for alanine substitution at residue 404 of antithrombin III Oslo suggests integrity of the 404-407 region is important for maintaining normal inhibitor levels. Thrombosis and Haemostasis 1989; 62: 494.
80. Nakagawa M, Tanaka S, Tsuji H, Takada O, Ono T. Congenital antithrombin deficiency (ATIII Kyoto): identification of a point mutation altering arginine-406 to methionine behind the reactive site. Thrombosis Research 1991; 64: 101-108.
81. Bock SC, Marrinan JA, Radziejewska E. Antithrombin III Utah: proline 407 to leucine mutation in a highly conserved region near the inhibitor reactive site. Biochemistry 1988; 27: 6171-6178.
82. Olds RJ, Lane DA, Caso R, Panico M, Morris HR, Sas G, Dawes J, Thein SL. Antithrombin III Budapest: a single amino acid substitution (429Pro to Leu) in a region highly conserved in the serpin super family. Blood 1992; 79: 1206-1212.
83. Bock SC, Levitan DJ. Characterisation of an unusual length polymorphism 5' to the antithrombin III gene. Nucleic Acids Research 1983; 11: 8569-8582.
84. Bock SC, Radziejewska E. A Nhe 1 RFLP in the human antithrombin III gene (1q23-25) (AT3). Nucleic Acids Research 1991; 19: 2519.
85. Daly ME, Perry DJ. Dde I polymorphism in intron 5 in the antithrombin III gene. Nucleic Acid Research 1990; 18: 5583.
86. Wu S, Seino S, Bell GI. Human antithrombin III (AT3) gene length polymorphism revealed by the polymerase chain reaction. Nucleic Acids Research 1989; 17: 6433.
87. Whisstock J, Skinner R, Lesk AM. An atlas of serpin conformations. TIBS 1998; 63-67
88. Desai UR, Petitou M, Bjork I, Olson ST. Mechanism of heparin activation of antithrombin. Journal of Biological Chemistry 1998; 273: 7478-7487.
89. Huntington JA, Gettins PGW. Conformational conversion of antithrombin to a fully activated substrate of factor Xa without need for heparin. Biochemistry 1998; 37: 3272-3277.
90. Jin L, Abrahams JP, Skinner R, Petitou M, Pike RN, Carrell RW. The anticoagulant activation of antithrombin by heparin. Proceedings of the National Academy of Sciences USA 1997; 94: 14683-14688.
91. Lane DA, Ireland H, Olds RJ, Thein SL, Perry DJ, Aiach M. Antithrombin III: a database of mutations.Thrombosis and Haemostasis 1991; 66: 657-661.
92. Lane DA, Olds RJ, Boisclair M, Chowdhury V, Thein SL, Cooper DN, Blajchman M, Perry D, Emmerich J, Aiach M. Antithrombin III mutation database: first update. For the Thrombin and its Inhibitors Subcommittee of the Scientific and Standardization Committee of the International Society on Thrombosis and Haemostasis. Thrombosis and Haemostasis 1993; 70: 361-369.
93. Lane DA, Bayston T, Olds RJ, Fitches AC, Cooper DN, Millar DS, Jochmans K, Perry DJ, Okajima K, Thein SL, Emmerich, J. Antithrombin mutation database: 2nd (1997) update. Thrombosis and Haemostasis 1997; 77: 197-211.
94. Antonarakis SE, Nomenclature working group. Recommendations for a nomenclature system for human gene mutations. Human Mutation 1998; 11: 1-3.
95. Olds et al, Unpublished
96. Emmerich et al, Unpublished
97. Jochmans et al, Unpublished
98. Perry et al, Unpublished
99. Blajchman et al Unpublished
100. Okajima et al, Unpublished
101. Aiach et al, Unpublished
102. Daly et al, Unpublished
103. Keeling et al Unpublished
-
Compiled by members of the Plasma Coagulation Inhibitors Subcommittee of the Scientific and Standardization Committee of the International Society on Thrombosis and Haemostasis.
- The final version of the database was put together by Trevor Bayston and David Lane (Centre for Haematology, Imperial College London).
- Image credit - Animated antithrombin molecule: this image is derived from the co-ordinates of Professor Robin Carrell and his group at the University of Cambridge (UK).
Note to database users
Please note that this database is no longer updated.